Splitting and Cleavage¶
With the Cluster concept previously introduced, we can do two very interesting things with them: Splitting and Cleavage. We define a split, we start with a cluster, and split it into two clusters, the original cluster, and a new daughter cluster, and we proportion the contents of the original cluster between itself, and the new dauger cluster. Cleavage on the other hand occurs in-place in a cluster, and divides the contents of the cluster, and creates new clusters contained within the orignal one.
The split operation is typically used to apply a cell division process, where single cell splits into itself, and a new daughter. The cleavalge operation models the early embryo cleavage processes.
Splitting¶
The Cluster.split() method splits a given cluster into itself, and a new
daughter cluster. Split accepts optional normal and point arguments to define a
cleavage plane. If only a normal vector is given, and no point, the split
uses the center of mass of the cluster as the point. For example, to split a
cluster along the x axis, we can:
c = MyCell(...)
d = c.split(normal=[1., 0., 0.])
where d is the new daughter cell. This form uses the center of mass of the cluster as the cleavage plane point. We can also specify the full normal/point form as:
d = c.split(normal=[x, y, z], point=[px, py, pz])
If no named arguments are given, split interprets the first argument as a cleavage plane normal:
c.split([x, y, z])
We frequently want to split a cell along an axis, i.e. generate a cleavage plane coincident with an axis. In this case, we use the optional axis argument. Here, the split will generate a cleavage plane that contains the given axis:
c.split(axis=[x, y, z])
The default version of split uses a random cleavage plane that intersects the cell center:
d = c.split()
Split can also randomly pick half the particles in a cluster, and assign them to a new cluster with the random agument as in:
c.split(random=True)
Cleavage¶
The Cluster.cleave() method is the single entry point for all cleavage
operations, and it supports a lot of different options.
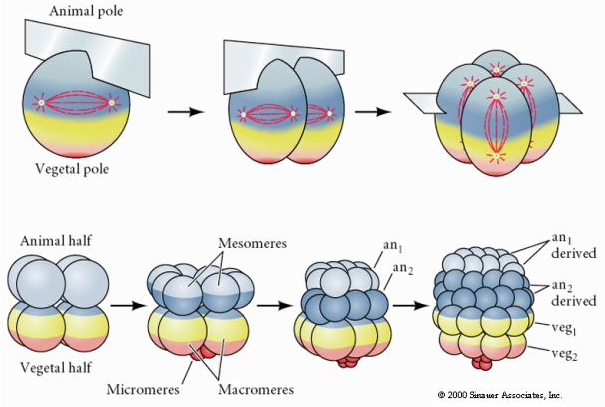
Cleavage is built on the concept of embryo cleavage. Embryology introduced some rather interesting terminolgy that we adopt here. First off, embryologists have a concept called an animal and vegatble poles. We represent these concepts using the built-in base Particle.orientation vector, and here we use this vector to represent a direction from the vegatable pole to the animal pole. All of the forms of the Cluster.cleave method will use the cluster’s orientation vector in this way. So, setting a cluster’s orientation vector, if the object is used to represent an embryo sets the direction of the pole. All of the contined cells can then refer to this property to get their orientation, i.e.:
embryo = MyEmbryo(orientation=[1.0, 0.0, 0.0], ...)
creates an new embryo with it’s pole aligned along the positive x axis.
The Cluster.cleave method is designed to perform the kinds of operations in:
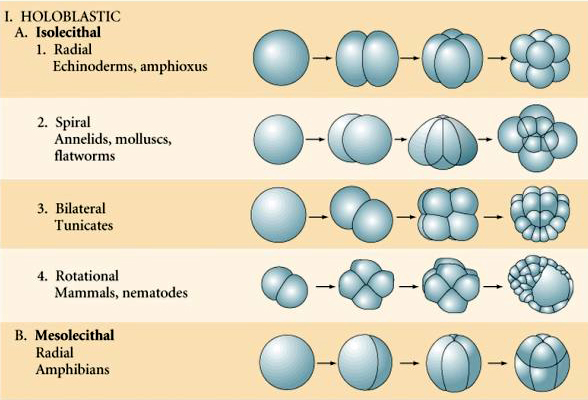
Radial Cleavage¶
We perform a radial cleavage using the kind=RADIAL argument, and a number that specifies what level of cleavage to perform. As in the above figure, level 1 splits a single cluster into two, along the orientation axis, level 2 splits it into 4 new clusters, orientated along the orietation axis, level 3 splits it into 8 new clusters, usign a cleavage plane as in
For example, if we create an embryo, and call:
embryo = MyEmbryo(orientation=[1.0, 0.0, 0.0], ...)
embryo.cleave(kind=RADIAL, level=3, cell_type=MyEmbryo.Basic)
this creates eight new clsuters inside the parent embryo cluster, and assigns all of them the MyEmbryo.Basic type. Or optionallly, we may use any Cluster derived type. If the cell_type option is left blank, the new cluster default to the top level Cluster type.
Subsequent Radial Cleavage¶
We don’t have to perform the cleavage in a single step, rather we can call:
embryo.cleave(kind=RADIAL, cell_type=MyEmbryo.Basic)
without specifing a level, in this case, ‘cleave` looks at the current number of contained clusters, and perfoerms the next pre-defined cleavage operations. For example, if the embryo only contained a single cell, then it pefrorms the first cleavage operation. If there are two cells, it splits them along the orientation axis, if there are 4, then it splits them perpendicular to the orientation axis, as in the previous figure.