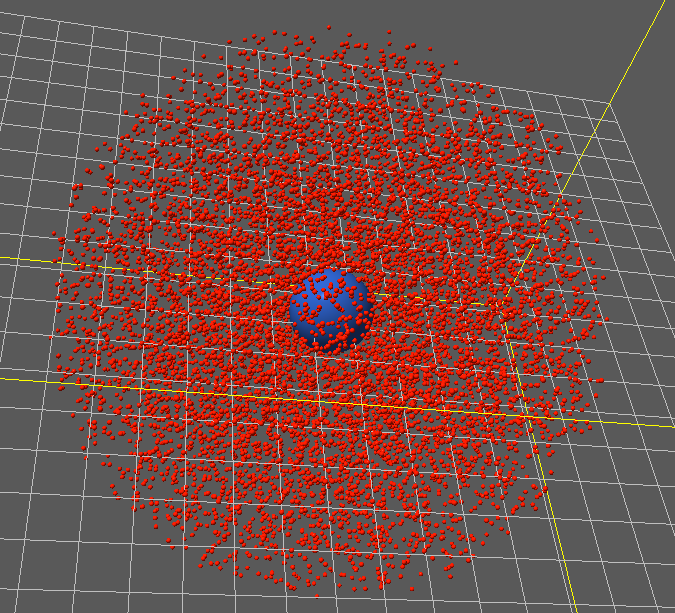
Cytoplasmic Particles¶
The motion of particles inside a cell is a very interesting scientific
problem. We have a number of ways of representing particles bound to a region,
we introduce the simplest method here. Mechanica provides a well
potential that creates a hard potential wall, and constrains particles inside
it. We can use the square well potential to model receptors in a cell’s
cytoplasm.
Start with the usual model setup:
import mechanica as m
import numpy as np
# potential cutoff distance
cutoff = 9
receptor_count = 10000
# dimensions of universe
dim=np.array([20., 20., 20.])
center = dim / 2
# new simulator, don't load any example
m.Simulator(dim=dim, cutoff=cutoff, cells=[4, 4, 4], threads=8)
Make a couple particle types, one to represent a nucleus, and another for receptors. We want the nucleus fixed in space, so make it very heavy:
class Nucleus(m.Particle):
mass = 500000
radius = 1
class Receptor(m.Particle):
mass = 0.2
radius = 0.05
target_temperature=1
#dynamics = m.Overdamped
Create a array of random positions within a solid sphere for the initial receptor positions:
# locations of initial receptor positions
receptor_pts = m.random_point(m.SolidSphere, receptor_count) * 5 + center
Make a well potential, and a soft_sphere potential. The well
confines the receptors to within a fixed distance of the nucleus, and the soft
sphere potential gives the receptors a definite shape, and prevents overlapping:
pot_nr = m.Potential.well(k=15, n=3, r0=7)
pot_rr = m.Potential.soft_sphere(kappa=15, epsilon=0, r0=0.3, eta=2, tol=0.05, min=0.01, max=1)
# bind the potential with the *TYPES* of the particles
m.bind(pot_rr, Receptor, Receptor)
m.bind(pot_nr, Nucleus, Receptor)
Create a random force (Brownian motion), zero mean of given amplitude, this gives the receptors some random motion.
tstat = m.forces.random(0, 3)
# bind it just like any other force
m.bind(tstat, Receptor)
Position the nucleus at the middle, and loop over the receptor points, and make a receptor, and finally run the simulation:
n=Nucleus(position=center, velocity=[0., 0., 0.])
for p in receptor_pts:
Receptor(p)
# run the simulator interactive
m.Simulator.run()
The complete simulation script is here, and can be downloaded here:
Download: this example script